Seek Ones Single Molecule Technologies
Our Technology
Handling single molecules with precision

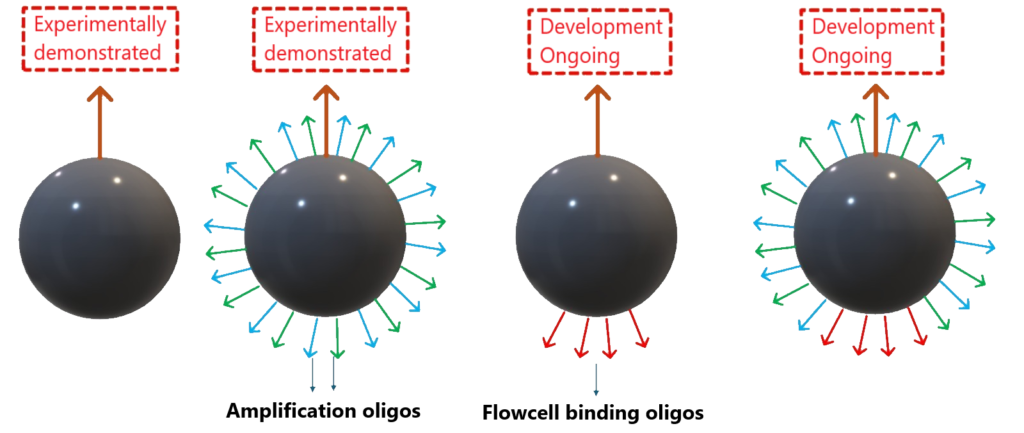
We have devised a patent-pending procedure to mass produce nanoparticles, each carrying exactly one capture moiety (see schematic). These can be exposed to analyte molecules, which will bind 1:1 to these capture moieties. This neutralizes Poisson (pun intended!) limitations, enabling high monoclonality and occupancy.
Further refinements can include amplification oligos, as well as flowcell binding oligos on the opposite pole from the capture moiety.
Applications
Application 1 :
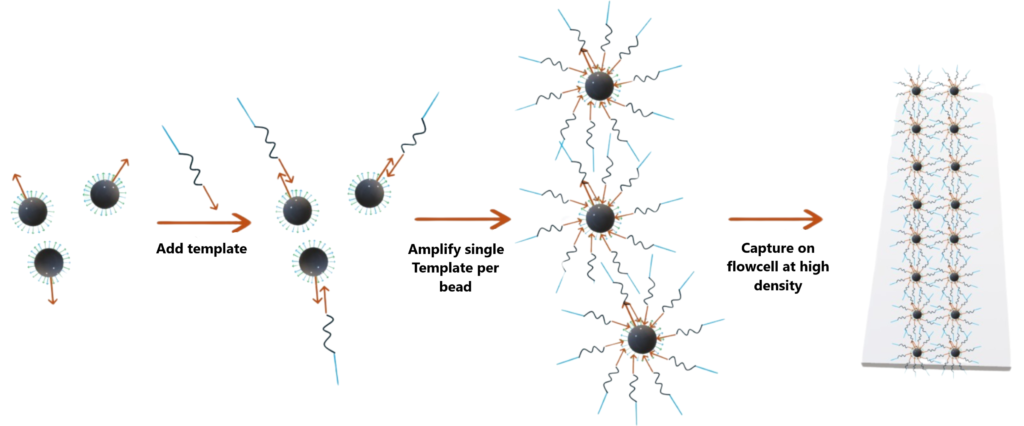
These beads can be used to capture DNA templates at a 1:1 ratio, followed by amplification and placement on a flowcell. By using beads with diameters of the desired resolution, we can pack in beads at high density without “overclustering” because the beads will maintain a fixed distance due to steric clashes.
Application 2 :
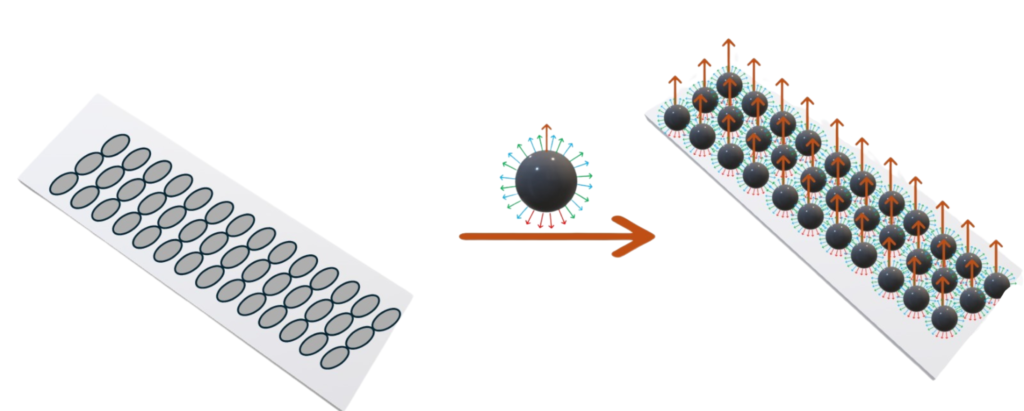
Single capture moiety beads can be pre-bound 1:1 to nanowells on a flowcell. When template are added, this will enable a single template to be captured per nanowell, improving both occupancy and monoclonality.
About us

I have 15 years experience in protein engineering and DNA sequencing and am trying to bring some ideas of my own to life. So far this is a solo effort but am looking for collaborative synergies. Drop me a line if you’re interested!
Contact Us
Email saurabh@seek1s.com for more information.
Or fill out the form below